from sklearn.metrics import accuracy_score
y_pred = [0, 2, 1, 3]
y_true = [0, 1, 2, 3]
accuracy_score(y_true, y_pred)0.5Accuracy is a metric for evaluating the performance of a classification algorithm. It can be understood as the fraction of predictions the model performed correctly.
\displaystyle \text{Accuracy} \ =\ \frac{\text{No of correct predictions}}{\text{Total number of predictions}}
\displaystyle \text{Accuracy} \ =\ \frac{\text{TP + TN}}{\text{TP + FP +TN + FN}}
Where, TP = True Positives FP = False Positives TN = True Negatives FN = False Negatives
Consider a dataset on 100 individuals where we would like to predict if an individual is suffering from cancer.
The dataset contains 90 individuals who do not suffer from cancer (class 0) whereas 10 of them who do (class 1). If we build a model that always predicts class 0, then our model will never be able to predict that an individual might be suffering from cancer. So, we establish that this is a bad model.
However based on the definition of accuracy, \displaystyle \text{Accuracy} \ =\ \frac{90}{100} = 0.9
From this, we can see that we would have to use other metrics when we have imbalanced datasets
link: https://scikit-learn.org/stable/modules/generated/sklearn.metrics.accuracy_score.html#sklearn.metrics.accuracy_score
To calculate accuracy using sklearn, we use the accuracy_score function from the module sklearn.metrics
The parameters are as follows:
from sklearn.metrics import accuracy_score
y_pred = [0, 2, 1, 3]
y_true = [0, 1, 2, 3]
accuracy_score(y_true, y_pred)0.5#normalize=False gives the count of correctly classified samples
accuracy_score(y_true, y_pred, normalize=False)2#Sample weights can be varied for each point
accuracy_score(y_true, y_pred, sample_weight=[0,0,0,1])1.0#To view a simplified version of the documentation in colab environment
accuracy_score?We are going to calculate the accuracy of a KNN classifier model on the breast_cancer dataset using sklearn.
The dataset does not have any categorical variables and we will just split the dataset into train and test. We shall fit the model on the training dataset and find the accuracy on the test dataset.
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
X,y = load_breast_cancer(return_X_y=True,as_frame=True)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=42)from sklearn.neighbors import KNeighborsClassifier
model = KNeighborsClassifier()
model.fit(X_train,y_train)
model.score(X_test,y_test)0.965034965034965y_pred = model.predict(X_test)
accuracy_score(y_test,y_pred)0.965034965034965from sklearn.metrics import ConfusionMatrixDisplay
ConfusionMatrixDisplay.from_estimator(model, X_test, y_test)<sklearn.metrics._plot.confusion_matrix.ConfusionMatrixDisplay at 0x7d7a6fb661a0>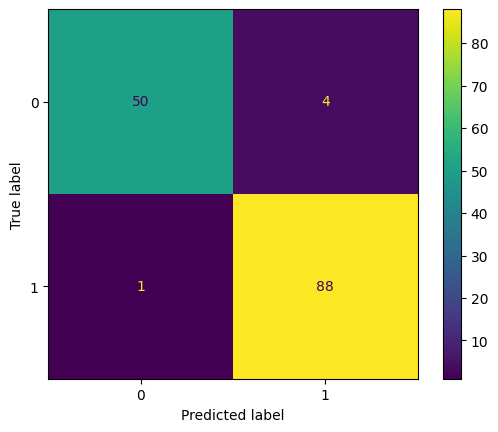
In the above matrix, we can see that: TP = 88, TN = 50, FP = 4, FN = 1
\begin{aligned} {\displaystyle \text{Accuracy}} & ={\displaystyle \frac{\text{TP + TN}}{\text{TP + FP +TN + FN}}}\\ & \\ & {\displaystyle =\frac{\text{88 + 50}}{\text{88 + 4 + 50 + 1}}}\\ & \\ & {\displaystyle =\ \frac{\text{138}}{\text{143}}}\\ & \\ & {\displaystyle =\ 0.965} \end{aligned}We use GridSearchCV for hyperparameter tuning and cross validation. We can change the scoring metric to tune the model as per our requirement.
from sklearn.model_selection import GridSearchCV
param_grid = {'n_neighbors':[3,5,10,20]}
model_tuned = GridSearchCV(estimator=KNeighborsClassifier(),param_grid=param_grid,scoring='accuracy')
model_tuned.fit(X_train,y_train)
model_tuned.score(X_test,y_test)0.972027972027972